structures that
contain both proteins and nucleic acids. The nucleic acid is most likely RNA because of its fibrous and decondensed nature as visualized by electron microscopy. The structural integrity of SNBs is influenced by transcription as transcriptional inhibitors and entry into mitosis dissembles SNBs (Chen et al., 1999)  .
.
|
SNB components
In addition to Sam68, two related Sam68 family members, Sam68 Like Mammalian (SLM)- 1 and SLM-2 (also known as T-Star or Etoile), have been shown to localize in SNBs (Chen et al., 1999)  . Other proteins found to localise to SNBs include: the alternative splicing factor YT521-B (Hartmann et al., 1999) . Other proteins found to localise to SNBs include: the alternative splicing factor YT521-B (Hartmann et al., 1999) ; the soluble tyrosine kinase BRK/SIK (Breast tumor Kinase/ Src-related Intestinal Kinase) (Derry et al., 2000) ; the soluble tyrosine kinase BRK/SIK (Breast tumor Kinase/ Src-related Intestinal Kinase) (Derry et al., 2000) ; and HAP (hnRNP A1 associated protein) (Denegri et al., 2001) ; and HAP (hnRNP A1 associated protein) (Denegri et al., 2001) . Interestingly, the SNBs are able to recruit some splicing factors of the SR family upon stress induction (Denegri et al., 2001) . Interestingly, the SNBs are able to recruit some splicing factors of the SR family upon stress induction (Denegri et al., 2001) . .
SNB function
The function of SNBs is unknown. SNBs have only been found in immortalized and primary cell lines. Thus, their predominant presence in highly transformed and poorly differentiated cell lines, such as HeLa and BT-20, suggest that SNBs may be a marker for these types of cancer cells. Many SLM proteins share SH2 and SH3 domains, suggesting this family of proteins may serve as adaptor proteins for cellular protein kinases. In addition, their ability to bind RNA and the presence of splicing factors in SNBs suggest that they may play a role in coupling signal transduction pathways to pre-mRNA processing. Further support for a role in RNA metabolism comes from the recent evidence that rat SLM-2 can interact via two-hybrid with several splicing factors (SRp30c, YT521-B and SAF B) and can regulate the alternative splicing of a CD44 mini gene (Stoss et al., 2001) . .
| 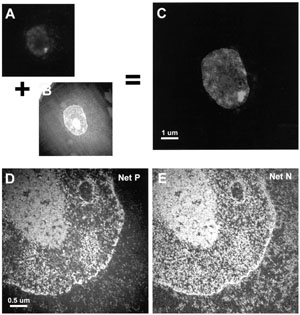
Transmission EM Image of a Sam68 Body
A) Immunfluorescence image of a Sam68 body within a Hela cell, which is combined with B) a transmission electron micrograph (TEM) to give a C) composite IF and TEM image. D) Net phosphorous (P) content using spectroscopic EM, indicates total nucleic acid content and E) Net nitrogen content (N) indicates total protein content of this cell. Note that Sam68 bodies contain both protein and nucleic acid.
Click to enlarge
(Courtesy: F-M. Boisvert)
|
Sam68/SLM Research from Dr. Stephane Richard (McGill University)
IF of Sam68/SLM bodies in HeLa cells from F.-M. Boisvert
REFERENCES
Chen, T., Boisvert, F.M., Bazett-Jones, D.P. and Richard, S. (1999) A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell 10 (9):3015-33.
Denegri, M., Chiodi, I., Corioni, M., Cobianchi, F., Riva, S. and Biamonti, G. (2001) Stress-induced Nuclear Bodies Are Sites of Accumulation of Pre-mRNA Processing Factors. Mol. Biol. Cell 12 (11):3502-3514.
Derry, J.J., Richard, S., Valderrama Carvajal, H., Ye, X., Vasioukhin, V., Cochrane, A.W., Chen, T. and Tyner, A.L. (2000) Sik (BRK) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol. Cell. Biol. 20 (16):6114-26.
Hartmann, A.M., Nayler, O., Schwaiger, F.W., Obermeier, A. and Stamm, S.(1999) The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol. Biol. Cell 10 (11):3909-26.
Huang S. (2000) Review: perinucleolar structures. J. Struct. Biol. 129 (2-3): 233-40.
Stoss O, Olbrich M, Hartmann AM, Konig H, Memmott J, Andreadis A, Stamm S. (2001) The STAR/GSG family protein rSLM-2 regulates the selection of
alternative splice sites. J. Biol. Chem. 276(12):8665-8673